Claudia Ball Group
Experimental Translational Oncology (ETO)

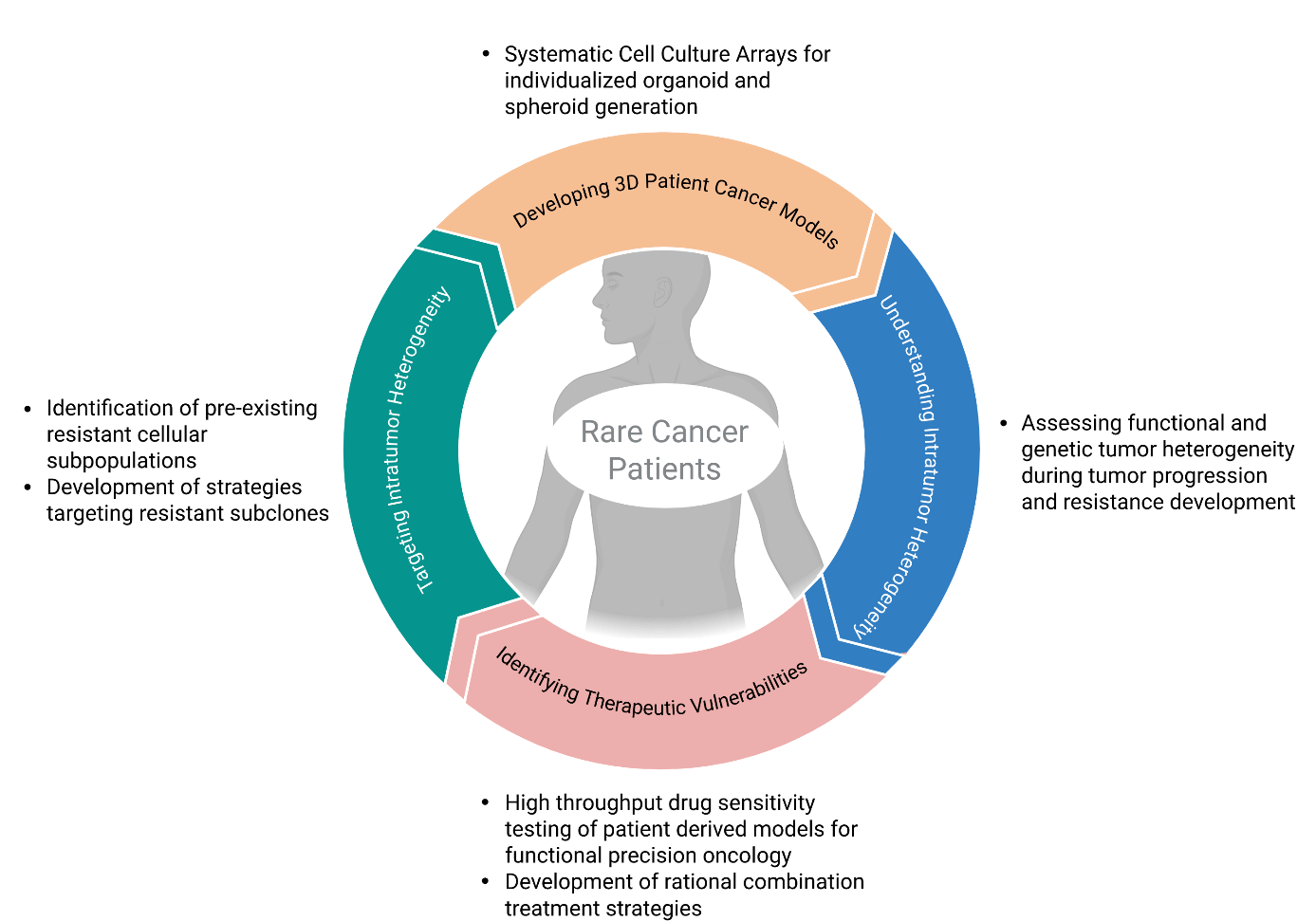
Claudia Ball leads the experimental translational oncology group at the National Center for Tumor Diseases (NCT/UCC) Dresden with a focus on functional precision oncology in rare and therapy-resistant cancers. Her team develops and applies patient-derived three-dimensional (3D) tumor models to functionally assess drug response and support personalized treatment decision-making. These efforts aim to complement and extend molecular diagnostics by adding a functional readout of tumor vulnerability, particularly in clinically underserved tumor entities.
The group has established a robust and scalable platform for ex vivo drug sensitivity testing, covering both short-term patient-derived cultures from fresh biopsies and long-term expandable 3D models such as spheroids and organoids. Using automated workflows and dose-response profiling of clinically relevant agents, the drug sensitivity test platform enables high-content phenotypic screening even with minimal starting material. All models are molecularly annotated and linked to patient-specific clinical and histopathological data, creating a resource to identify individual and shared therapeutic vulnerabilities. Drug sensitivity testing results are functionally integrated into a clinical decision-making pipeline within a precision oncology framework for rare cancers. In this context, the group collaborates closely with physician scientist in multidisciplinary tumor boards to inform treatment recommendations based on functional profiling. In addition, the team contributes to the development of tools for interpreting and prioritizing drug sensitivity data for clinical application.
Recent work has functionally validated DNA repair-associated biomarkers, such as the TOP-ART score, in patient-derived models and linked HRD status to specific patterns of sensitivity to PARP inhibitors and DNA-damaging agents. These findings support the refinement of stratification strategies beyond genomic annotation alone.
Beyond classical viability-based response profiling, the group is currently exploring additional layers of information for patient stratification, including label-free imaging as well as and metabolic fingerprinting, to resolve functional heterogeneity, assess drug responses on the single cell level and identify additional layers of vulnerability. These multimodal approaches aim to capture dynamic response patterns, clonal outgrowth, and non-genetic resistance mechanisms in heterogeneous tumor populations. Looking ahead, the group is expanding its efforts to explore the influence of additional biological factors, such as niche signals, on drug response. A particular interest lies in integrating more complex model systems and readouts to better capture inter-patient heterogeneity and guide rational therapy development in rare and in treatment resistant cancers.
Future Projects and Goals
Future work will focus on expanding functional precision oncology strategies in rare and therapy-resistant cancers. A key aim is to understand what drives therapy resistance at the single-cell level within heterogeneous tumor populations. This will involve the integration of single-cell molecular and sensitivity profiling to resolve functional subclones and stratify patients more precisely. In parallel, the group plans to develop more complex model systems incorporating stromal and immune components to better reflect in vivo response dynamics. Long-term goals include refining functional predictors of drug sensitivity and translating these insights into clinically actionable, model-based treatment strategies.
Methodological and Technical Expertise
- Patient-derived 3D patient-derived cancer model (PCM) generation and characterization (organoids, spheroids, short-term PCMs)
- Ex vivo drug sensitivity testing in miniaturized and automated formats
- Functional precision oncology data integration and interpretation
- Analysis of clonal dynamics, therapy response and resistance